emotion.biomachina.org
... powered by Situs
| Home |
| About |
| Database |
| Deposit Map |
| F.A.Q |
| Situs |
| Biomachina |
About | ||
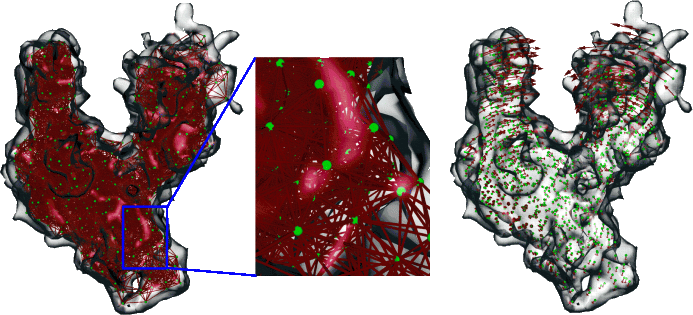
Introduction to Normal Mode Analysisemotion uses normal mode (or vibrational) analysis to characterize global motions of large-scale biomolecules. Traditionally, normal mode analysis (NMA) is applied to an atomic structure where the atomic interactions are described by a standard molecular dynamics force field. One of our main goals is to reduce the spatial resolution of elastic models of large biomolecular assemblies below the single residue level. This suggestion is rooted in the observation that atomic motions corresponding to low-frequency normal modes are not localized. Hence, it seems reasonable to expect that a sparse estimation of the displacement field using a spatial resolution of 5-10Å will reproduce the displacements well. After generating such a sparse estimation, displacements can be extended to full space by spline-interpolation. Moreover, a mesoscopic description of the dynamics can be applied directly to 3D image reconstructions from EM, as it is independent of the resolution of the underlying data. Consequently, we use the landmarks that can be computed with our vector quantization algorithm implemented in the Situs package to create a deformable model.As an example, we demonstrate how essential motions can be extracted from a low-resolution EM map in the absence of a known atomic structure:
It is important to reiterate that NMA is based on a harmonic model of the displacements, as if the molecule was moving as an elastic body. The returned normal modes are then ordered by their frequency and generally the lowest-frequency modes are observed to be functionally most relevant. For more info on the interpretation of our emotion animations please read our F.A.Q. page! | ||
Reference: | ||
|
Pablo Chacon, Florence Tama and Willy Wriggers. Mega-Dalton Biomolecular Motion Captured from Electron Microscopy Reconstructions. J. Mol. Biol., 326:485-492, 2003. [Abstract] [Article] |
Last Modified: